Escherichia coli uropatogenă şi factorii de virulenţă înrudiţi
Uropathogenic Escherichia coli and the related virulence factors
Abstract
Uropathogenic strains of Escherichia coli (UPEC) are the most common cause of non-hospital-acquired urinary tract infections (UTIs). The most common UTIs occur mainly in women and affect the bladder and urethra, leading to infections of the bladder (cystitis). Uropathogenic Escherichia coli have genes and virulence factors in association with adhesion, biofilm formation and colonization. These genes are: mrk, kpsM, foc, auf, C, Kps, chuA, hma, ireA, iha, iutA, fliC, ompA, upab, upaC, upaG, and upaH. Doctors typically treat UTIs with a wide range of different antibiotics, such as fosfomycin, nitrofurantonin, pivmecillinam, trimethoprim, sulfamethoxazole, ciprofloxacin, levofloxacin and prulifloxacin. However, some strains of E. coli, called extended-spectrum beta-lactamase (ESBL) E. coli, are resistant to most antibiotic treatments.Keywords
urinary tract infectionsEscherichia coliESBLRezumat
Tulpinile uropatogene de Escherichia coli sunt cea mai frecventă cauză de infecţii non-intraspitaliceşti ale tractului urinar. Infecţiile de tract urinar apar mai frecvent la femei şi afectează vezica urinară şi uretra. Infecţia vezicii urinare poartă denumirea de cistită. Escherichia coli uropatogen prezintă factori de virulenţă şi gene responsabile de aderare, formarea biofilmului şi colonizare. Principalele gene sunt: mrk, kpsM, foc, auf, C, Kps, chuA, hma, ireA, iha, iutA, fliC, ompA, upab, upaC, upaG şi upaH. Pentru tratarea infecţiei, medicii apelează în mod obişnuit la o gamă largă de antibiotice diferite, cum ar fi fosfomicină, nitrofurantonină, pivmecilinam, trimetoprim, sulfametoxazol, ciprofloxacină, levofloxacină şi prulifloxacină. Cu toate acestea, unele tulpini de E. coli, denumite beta-lactamaze cu spectru extins (ESBL) E. coli, sunt rezistente la majoritatea tratamentelor cu antibiotice.Cuvinte Cheie
infecţii de tract urinarEscherichia coliESBLIntroduction
Despite many progresses in the field of medical microbiology, urinary tract infections (UTIs) are still known as a big concern for microbiologists and stay as the second ranking infectious diseases all over the world. The most important problem with the UTIs is related to their multimicrobial spectrum. Either bacterial or fungal agents involve lower part (cystitis) and/or upper part (pyelonephritis) of urinary tract. Escherichia coli strains and in particular the uropathogenic pathotypes are recognized as the most important bacterial etiology of the UTIs, while the fungal agent of Candida albicans has the same role; however, C. albicans may cause different types of candidiasis, including UTIs.
E. coli bacteria are divided into two groups of intra-intestinal and extra-intestinal strains. The intra-intestinal strains may be categorized into two subgroups of commensal strains and the pathotypes. On the other hand, the extra-intestinal strains include different types of pathotypes such as uropathogenic E. coli (UPEC). UPEC pathotypes are known as invaluable genomic treasures of a wide range of diverse virulence factors which each one has its own importance regarding UTIs. Antigens of Flagella (H), Soma (O) and Capsule (K) are common criteria for UPEC strains classification. In addition to varieties of virulence factors, the feature of antibiotic resistance in UPEC pathotypes is a big deal to be concerned for. The progression of multi-drug resistant (MDR) and extensively drug resistant (XDR) strains of UPEC worldwide has complicated the treatment of the UTIs, too(1-12).
As mentioned before, the treasure of virulence factors and genes within the pan-genome of UPEC strains is amazingly widespread. But, in this review, some of them which are involved in biofilm formation have been studied (Table 1 and Figure 1)(6,11-13).
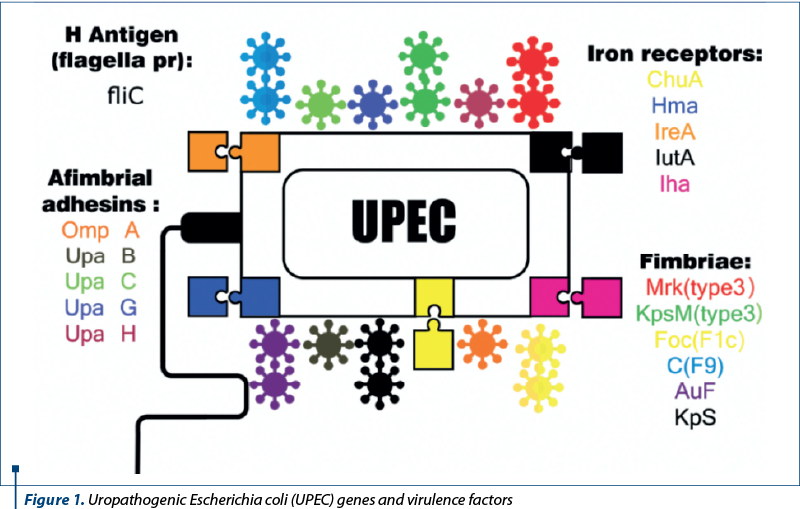
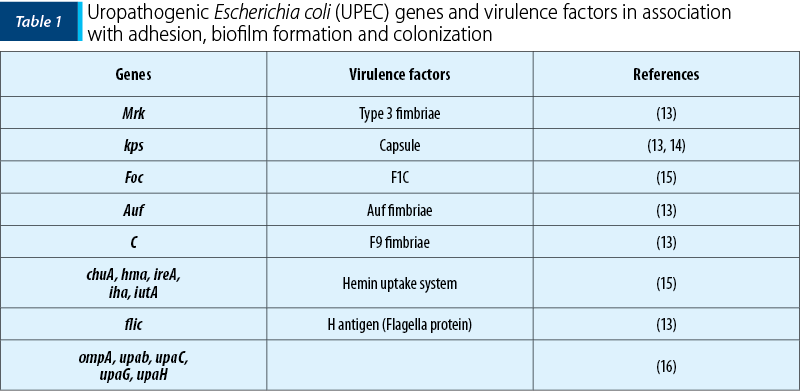
Materials and method
In silico studies have been done for this article. Databases such as NCBI and GenBank in parallel with Google Scholar were searched to find and study various review articles about UPEC genes from the year 2000 to 2018. We have also used reference books to improve our data.
Genes involved in biofilm formation
As indicated in Table 1, genes which have contributed in biofilm formation can be divided into two groups: biofilm formation in catheher and the host’s body. Some of the most important virulence genes of UPEC strains which are associated with biofilm formation are, mrk, kpsM, foc, auf, C, Kps, chuA, hma, ireA, iha, iutA, fliC, ompA, upab, upaC, upaG and upaH. Among them, mrk and kpsM can form biofilms on indwelling catheters and others in the host’s body. Specific virulent properties of each gene have been analyzed in this study.
1. mrk
Catheter-associated urinary tract infection (CAUTI) had been relatively abandoned in clinical research until recently. CAUTIs occur from the growth of bacterial biofilms on the inner surface of the urinary catheter. The urinary catheters are tubular latex or silicone devices, which when inserted, biofilms can be formed on the inner or outer surfaces(17). Biofilm formation is typically facilitated by fibrillar structures such as fimbriae or pili. In Escherichia coli, the production of determined types of fimbriae (e.g., type 3 fimbriae) increases biofilm formation. One of the genes that are deeply associated with biofilm formation has been named as mrk gene. The genetic structures in a cell which can replicate independently of the chromosomes are called plasmids. Plasmids are circular DNA in the cytoplasm of a bacterium. Mrk gene located in plasmid (pMAS2027) encodes the type 3 fimbriae. The precise position of mrk gene is on a segment (5,536-bp) with G+C content of 56.6%. It has been demonstrated that mrk genes are located on a mobile genetic element and on upstream and downstream of the mrk cluster; they are genes encoding proteins associated with transposition(18).
2. kpsM
Capsule (also known as K antigen) is a large structure of some prokaryotic cells such as bacteria. It is a polysaccharide layer that lies outside the cell envelope of bacteria, and is thus deemed part of the outer envelope of a bacterial cell. It is a layer which is not easily washed off, and it can be the cause of various diseases. Capsule is located immediately exterior to the peptidoglycan layer of Gram-positive bacteria and the Lipopolysaccharide layer of Gram-negative bacteria(19). In recent years, several studies have examined the role of polysaccharide capsules in the pathogenesis of UTI and polysaccharide capsules have been identified, and they have been divided into three groups(20). Remarkably, kpsM gene encodes a capsule transport protein which is one member of the gene cluster responsible for type 2 capsular polysaccharide synthesis. This gene is one of the reasons of biofilm formation. It is recognized that the deletion of the gene kpsM would cause a huge decrease in the virulence of the UPEC strain both in vitro and in vivo. Many studies show that kpsM was relatively highly conserved during the evolution process of bacterial species, so we can conclude that this gene is probably a molecular clock(21).
3. foc
Biofilm formation in bacteria including UPEC strains has determined increase survival in natural environments and in the host’s body(22). Foc gene encodes F1C fimbriae which is located on the bacterial surface and is capable of adhesion to mucosal and endothelial cells(23). F1C fimbriae are essential for biofilm formation on an inert surface. Among three genes encoding for fimbrial adhesive systems (fimH, pap and sfa/foc), the prevalence of sfa/foc gene had been found 23%(24). Many antibiotics have proven to be effective for the clinical symptoms of UTIs, but recurrent and chronic infections continue to afflict many people. Saira Bashir et al.(25) found that, due to indiscriminate use of drugs in developing countries, the pathogenic bacteria are more battle-hardened as compared with developed countries. Studies show that the resistance of nalidixic acid can be related to considerably decreased prevalence of the gene sfa/foc (S and F1C fimbriae)(26). Daniël J. Wurpel et al. observations along with the binding specificity of these organelles proved that F1C pili may impact the pathogenesis of a remarkable number of UTI cases. b-GalNac-1, 4b-Gal residues on glycolipids expressed by epithelial cells of the distal tubules and collecting ducts of the kidney and also by bladder and kidney endothelial cells. F1C pili are encoded by approximately 14% of UPEC isolates and can bind b-GalNac-1 and 4b-Gal residues(27).
4. auf
Buckles et al.(28) determined that auf gene cluster was significantly associated with uropathogenic E. coli isolates. The role of auf genes is in biofilm formation and adhesion. Auf fimbriae (encoded by auf gene) are encoded by 67% of UPEC strains and 27% fecal E. coli strains (commensal isolates)(29). Furthermore, auf can cause all types of UTIs. Each type of UTI may result in more-specific signs and symptoms, depending on which part of the urinary tract is infected. Acute pyelonephritis usually occurs with upper back and side (flank) pain, high fever, shaking and chills, nausea and vomiting signs(30). Cystitis can be recognized with signs and symptoms such as pelvic pressure, lower abdomen discomfort, frequent, painful urination, and blood in urine(31). Urethritis are often seen with burning with urination and discharge(32).
5. C
F9 Fimbriae of uropathogenic Escherichia coli mediates biofilm formation which encodes by C gene. F9 fimbriae expression was indicated at 20c, representing the first proof of functional F9 fimbriae expression by wild-type E. coli. However, for binding directly to the f9 promoter, the f9 gene expresses at 37c(27). Besides of UTIs, this fimbriae can be effective at the clinical symptoms of pyelonephritis. Pyelonephritis is a common infection in adult women, but there is a paucity of controlled trials of its treatment and the optimum duration of antibiotic treatment has not been properly defined(33). UPEC strains have different specific genomes, for example the UPEC strain CFT073 genome contains ten gene loci that share sequence identity with the chaperone-usher class of fimbriae. In this strain, the f9 has c number named c1931-c1936 and the genes are c1936-34-ydeSRQ, and these information in other strains can be various(34).
6. Kps
Biofilms are the microbial communities of the surface-attached cells which are embedded in a self-produced extracellular polymeric matrix. Kps genes encode K1, K2, K3, K5, K12, K13, K20 and K51/KspMT capsular polysaccharides. Among the K group, k1 and k2 are more outstanding. The K1 capsule on the surface of UPEC strains is a key virulence factor and its expression may be important in the beginning and development of UTIs and cystitis(35). Similar to other bacterial polysaccharide capsules, K1 capsule has two classical roles which contain inhibition of phagocytosis by granulocytes/monocytes and serum resistance. It has been recognized that the polysialic acid K1 capsule may not only protect UPEC from natural immunity, but also form an IBC matrix component which facilitates the aggregation of the bacterial communities, which in turn precludes infiltration of host inflammatory mediators and environmental stressors(36). While K1 played an insignificant role in conferring serum resistance, the K2 antigen is mildly important. In the presence of whole blood, both K1 and K2 antigens provided a survival advantage to the UPEC strains tested(37).
7. chuA, hma, ireA, iha, iutA
UPEC, the famous cause of urinary tract infection, uses specific outer membrane receptors, facilitating the import of iron-chelating siderophores and iron from host cells. For the colonization of the urinary tract by uropathogenic Escherichia coli, iron attainment mediated by special outer membrane receptors is essential. Heme is an essential source of iron for UPEC in the kidney. Siderophores are small, iron-chelating molecules which bind and transport iron in microorganisms. Many distinguished types of siderophores are produced by bacteria. For the synthesis of a siderophore after many steps in E. coli, the completed molecules are discharged instantly from the cytoplasm to the extracellular space through a complex named TolC. ChuA, hma, ireA, iha, and iutA genes work together for UPEC iron uptake system and chuA performs the task of heme transport. The transcriptional regulators influence on the expression of iron uptake genes. The expression of chuA is also regulated by RfaH(38). Iha is an abbreviation of the IrgA homolog adhesin which transport both enterobactin and dihydroxy benzoylserine (DHBS) and also help UPEC to fitness in the urinary tract(39).
8. filC
The structure of the flagellum is free at one end and attached to the cell at the other end. The bacterial flagella with a diameter of about 20 nm move the bacteria towards nutrients and other attractants. The flagellated Escherichia coli spp. are common causes of urinary tract infections and the flagella help the bacteria by propelling up the urethra into the bladder. There are four types of flagellar arrangement: monotrichous, amphitrichous, lophotrichous and peritrichous(40). FliC gene encodes the H antigen or flagella protein. The expression of fliC gene in vitro is optimal and increasing osmolarity combined with lowering pH represses fliC activity which is the similar condition to the environment of the bladder(41). According to a study by M.R. Karam Asadi et al.(42), about 70% of clinical isolates obtained from patients with UTI harbored fliC gene, so we conclude that fliC is another conserved gene among UPEC strains.
9. ompA, upab, upaC, upaG, upaH
The pathogenic roles of OmpA proteins, including adhesion, invasion or intracellular survival, have been expected. Features of the outer membrane protein A (OmpA) encoded by ompA gene are monomeric, main, integral, porin and heat-modifiable component of the bacteria. The ompA functions as an intracellular virulence for UPEC and also within bladder epithelium it causes persistent infection. It has been demonstrated that the deletion of the ompA gene did not disrupt the initial epithelial binding and invasion by UPEC, whereas it did preclude completion of the intracellular bacterial community (IBC) pathway(43).
The formation of the extracellular matrix (ECM) requires cells to secrete ECM proteins(44). UpaB is located at the bacterial cell surface. The function of upaB is cell adhesion and upaB can mediate the adherence to several ECM proteins, so the deletion of upaB can reduce the early colonization of the bladder.
The upaB gene is common among UPEC strains and is present in all available UPEC genomes, but absent or disrupted in all diarrheagenic E. coli genomes(45). UpaC AT-encoding gene is common in E. coli. Autotransporter (AT) proteins have their independent transport across the bacterial membrane system and final routing to the cell surface which facilitates by special structural properties. Several AT proteins have been characterized from UPEC, and these include the upaC protein which encodes by upaC gene(46). UpaG is a member of the trimeric autotransporter family of adhesins in UPEC and is strongly associated with other E. coli strains. UpaG proteins encoded by upaG genes mediates adhesion to human bladder epithelial cells(47). UpaH is a identified autotransporter protein that contributes to biofilm formation and bladder colonization by UPEC. UpaH encodes a large cell surface-located AT protein that contributes to biofilm formation(48). As we mentioned before, ompA, upaB, upaC, upaG, and upaH genes encode various proteins, such as ag43, upaB, upaC, upaG and upaH proteins, with biofilm formation, adhesion and chronic infection tasks, causing chronic UTIs.
Effective antibiotics on UPEC
For the treatment of urinary tract infections, the first and second lines of the treatment can be used in a variety of antibiotics, for example: fosfomycin, nitrofurantonin, pivmecillinam, trimethoprim, sulfamethoxazole, ciprofloxacin, levofloxacin, and prulifloxacin(49).
It is noteworthy that ciprofloxacin, levofloxacin and prulifloxacin should not be used as the first line of treatment due to their high side effects(49).
Conclusions
Urinary tract infections are demonstrated to be one of the most controversial infections all over the world. The commonest cause of UTI is uropathogenic Escherichia coli, with different virulence factors encoding by virulence genes, such as mrk (encoding type 3 fimbriae), kpsM (encoding type 3 fimbriae), foc (encoding F1C), auf (Auf fimbriae), C (F9 fimbriae), Kps (encoding K antigen group), chuA, hma, ireA, iha, iutA (encoding Hemin uptake system), fliC (encoding H antigen), and ompA, upaB, upaC, upaG and upaH. Among these virulence factors, genes by encoding make the course of treatment harder.
More studies are also need to find the most appropriate treatment for this issue, but we searched and found some antibiotics group for the treatment, such as: fosfomycin, nitrofurantonin, pivmecillinam, trimethoprim, sulfamethoxazole, ciprofloxacin, levofloxacin and prulifloxacin.
Conflict of interests: The authors declare no conflict of interests.
Bibliografie
- Ochoa SA, Cruz-Córdova A, Luna-Pineda VM, Reyes-Grajeda JP, Cázares-Domínguez V, Escalona G, et al. Multidrug-and extensively drug-resistant uropathogenic Escherichia coli clinical strains: phylogenetic groups widely associated with integrons maintain high genetic diversity. Frontiers in Microbiology. 2016; 7:2042.
- Momtaz H, Karimian A, Madani M, Dehkordi FS, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Annals of Clinical Microbiology and Antimicrobials. 2013; 12(1):8.
- Behzadi P, Behzadi E. The microbial agents of urinary tract infections at central laboratory of Dr. Shariati Hospital, Tehran, Iran. Turk Klin Tip Bilim. 2008; 28(4):445.
- Behzadi P, Behzadi E, Ranjbar R. Urinary tract infections and Candida albicans. Central European Journal of Urology. 2015; 68(1):96.
- Behzadi P, Behzadi E. A study on apoptosis inducing effects of UVB irradiation in Pseudomonas aeruginosa. Roum Arch Microbiol Immunol. 2011; 70:74-7.
- Behzadi P, Najafi A, Behzadi E, Ranjbar R. Microarray long oligo probe designing for Escherichia coli: an in-silico DNA marker extraction. Central European Journal of Urology. 2016; 69(1):105.
- Behzadi E, Behzadi P. The role of toll-like receptors (TLRs) in urinary tract infections (UTIs). Central European Journal of Urology. 2016; 69(4):404.
- Ranjbar R, Tabatabaee A, Behzadi P, Kheiri R. Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) genotyping of escherichia coli strains isolated from different animal stool specimens. Iranian Journal of Pathology. 2017; 12(1):25-34.
- Behzadi P, Behzadi E, editors. Uropathogenic Escherichia coli: An ideal resource for DNA microarray probe designing. International Conference on Bioinformatics and Biomedical Engineering; 2017: Springer.
- Behzadi P, Behzadi E. Evaluation of UVB light efficacy for inducing apoptosis in Candida albicans cultures. Roum Arch Microbiol Immunol. 2012; 71(1):39-42.
- Totsika M, Gomes Moriel D, Idris A, A Rogers B, J Wurpel D, Phan M-D, et al. Uropathogenic Escherichia coli mediated urinary tract infection. Current Drug Targets. 2012; 13(11):1386-99.
- Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrobial Resistance & Infection Control. 2016; 5 (1):11.
- Jahandeh N, Ranjbar R, Behzadi P, Behzadi E. Uropathogenic Escherichia coli virulence genes: invaluable approaches for designing DNA microarray probes. Central European Journal of Urology. 2015; 68(4):452.
- Donnenberg M. Escherichia coli: pathotypes and principles of pathogenesis. Academic Press; 2013.
- Pitout JD. Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Review of Anti-Infective Therapy. 2012; 10(10):1165-76.
- Agarwal J, Srivastava S, Singh M. Pathogenomics of uropathogenic Escherichia coli. Indian Journal of Medical Microbiology. 2012; 30(2):141.
- Tambyah PA, Oon J. Catheter-associated urinary tract infection. Current Opinion in Infectious Diseases. 2012; 25(4):365-70.
- Ong C-LY, Beatson SA, McEwan AG, Schembri MA. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Applied and Environmental Microbiology. 2009; 75(21):6783-91.
- Reid AN, Whitfield C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. Journal of Bacteriology. 2005; 187(15):5470-81.
- Buckles EL, Wang X, Lane MC, Lockatell CV, Johnson DE, Rasko DA, et al. Role of the K2 capsule in Escherichia coli urinary tract infection and serum resistance. The Journal of Infectious Diseases. 2009; 199(11):1689-97.
- Zong B, Liu W, Zhang Y, Wang X, Chen H, Tan C. Effect of kpsM on the virulence of porcine extraintestinal pathogenic Escherichia coli. FEMS Microbiology Letters. 2016; 363(21):fnw232.
- Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, et al. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Applied and Environmental Microbiology. 2009; 75(1):246-51.
- Emody L, Kerenyi M, Nagy G. Virulence factors of uropathogenic Escherichia coli. International Journal of Antimicrobial Agents. 2003; 22:29-33.
- Usein CR, Damian M, Tatu‐Chitoiu D, Capusa C, Fagaras R, Tudorache D, et al. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. Journal of Cellular and Molecular Medicine. 2001; 5(3):303-10.
- Bashir S, Sarwar Y, Ali A, Mohsin M, Saeed MA, Tariq A, et al. Multiple drug resistance patterns in various phylogenetic groups of uropathogenic E. coli isolated from Faisalabad region of Pakistan. Brazilian Journal of Microbiology. 2011; 42(4):1278-83.
- Horcajada JP, Soto S, Gajewski A, Smithson A, de Anta MTJ, Mensa J, et al. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. Journal of Clinical Microbiology. 2005; 43(6):2962-4.
- Wurpel DJ, Totsika M, Allsopp LP, Hartley-Tassell LE, Day CJ, Peters KM, et al. F9 fimbriae of uropathogenic Escherichia coli are expressed at low temperature and recognise Galβ1-3GlcNAc-containing glycans. PloS One. 2014; 9(3):e93177.
- Buckles EL, Bahrani-Mougeot FK, Molina A, Lockatell CV, Johnson DE, Drachenberg CB, et al. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infection and Immunity. 2004; 72(7):3890-901.
- Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HL. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of ygi and yad fimbriae. Infection and Immunity. 2011; 79(12):4753-63.
- Car J. Urinary tract infections in women: diagnosis and management in primary care. BMJ. 2006; 332(7533):94-7.
- Kim A, Lim B, Song M, Choo M-S. Pretreatment features to influence effectiveness of intravesical hyaluronic Acid instillation in refractory interstitial cystitis/painful bladder syndrome. International Neurourology Journal. 2014; 18(3):163.
- Brill JR. Diagnosis and treatment of urethritis in men. Am Fam Physician. 2010; 81(7):873-8.
- Sandberg T, Skoog G, Hermansson AB, Kahlmeter G, Kuylenstierna N, Lannergård A, et al. Ciprofloxacin for 7 days versus 14 days in women with acute pyelonephritis: a randomised, open-label and double-blind, placebo-controlled, non-inferiority trial. The Lancet. 2012; 380(9840):484-90.
- Ulett GC, Mabbett AN, Fung KC, Webb RI, Schembri MA. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology. 2007; 153(7):2321-31.
- King JE, Owaif HAA, Jia J, Roberts IS. Phenotypic heterogeneity in expression of the K1 polysaccharide capsule of uropathogenic Escherichia coli and downregulation of the capsule genes during growth in urine. Infection and Immunity. 2015;83(7):2605-13.
- Anderson GG, Goller CC, Justice S, Hultgren SJ, Seed PC. Polysaccharide capsule and sialic acid-mediated regulation promote biofilm-like intracellular bacterial communities during cystitis. Infection and Immunity. 2010; 78(3):963-75.
- Sarkar S. Investigating the virulence potential of the multidrug resistant uropathogenic Escherichia coli ST131 clone. PhD Thesis, 2013.
- Hagan EC. Iron acquisition by uropathogenic Escherichia coli: ChuA and Hma heme receptors as virulence determinants and vaccine targets: University of Michigan; 2009.
- Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HL, et al. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infection and Immunity. 2005; 73(2):965-71.
- Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. International Review of Cytology. 2004; 233:93-135.
- Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT. Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infection and Immunity. 2002; 70(3):1391-402.
- Asadi KM, Oloomi M, Habibi M, Bouzari S. Cloning of fimH and fliC and expression of the fusion protein FimH/FliC from Uropathogenic Escherichia coli (UPEC) isolated in Iran. Iranian Journal of Microbiology. 2012; 4(2):55.
- Nicholson TF, Watts KM, Hunstad DA. OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infection and Immunity. 2009; 77(12):5245-51.
- Sottile J, Hocking DC. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Molecular Biology of the Cell. 2002; 13(10):3546-59.
- Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, et al. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infection and Immunity. 2012; 80(1):321-32.
- Allsopp LP, Beloin C, Moriel DG, Totsika M, Ghigo J-M, Schembri MA. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli. Journal of Bacteriology. 2012; 194(21):5769-82.
- Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, et al. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. Journal of Bacteriology. 2008; 190(12):4147-61.
- Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, et al. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infection and Immunity. 2010; 78(4):1659-69.
- Bartoletti R, Cai T, Wagenlehner FM, Naber K, Johansen TE. Treatment of urinary tract infections and antibiotic stewardship. European Urology Supplements. 2016 Jul 1; 15(4):81-7.


